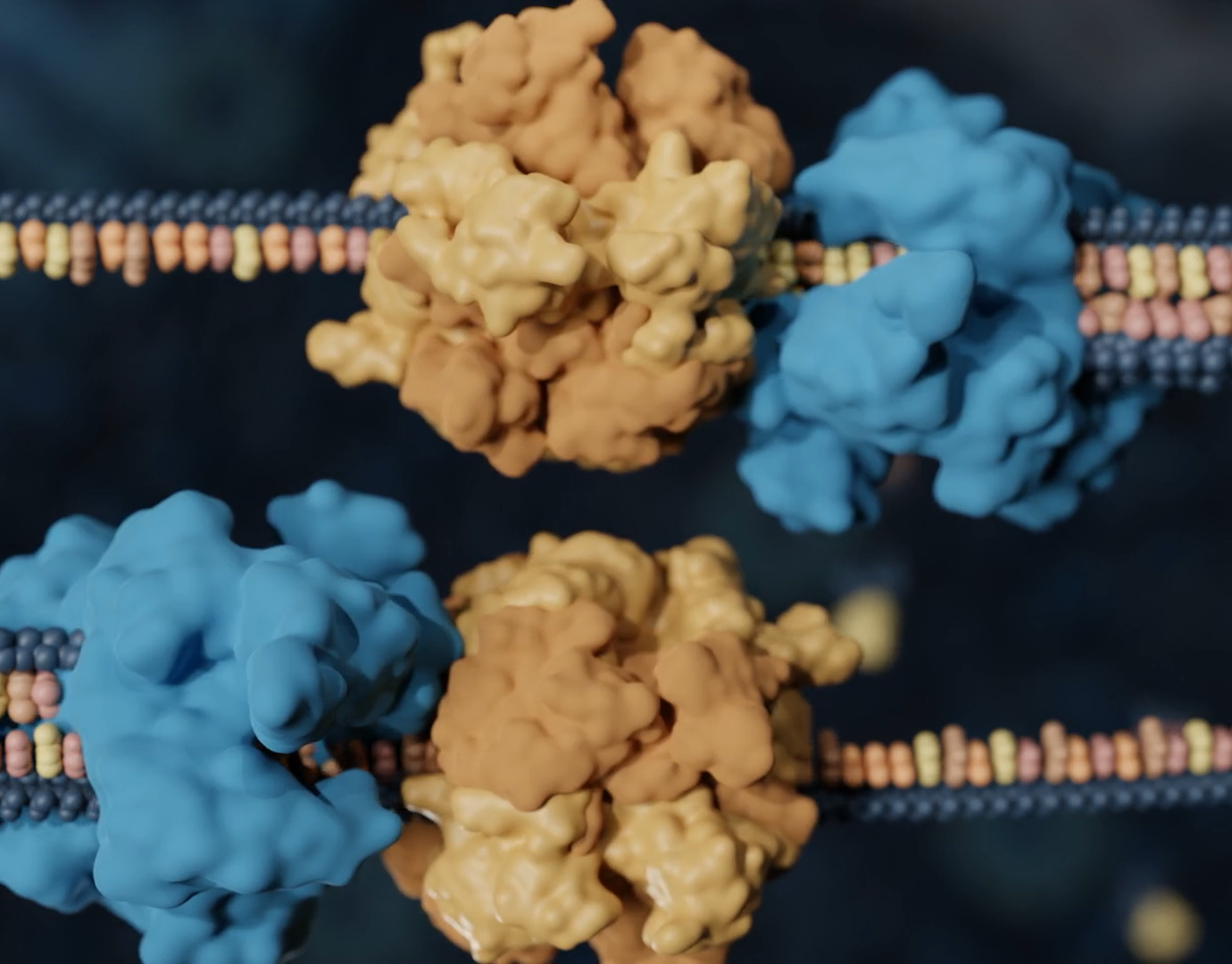
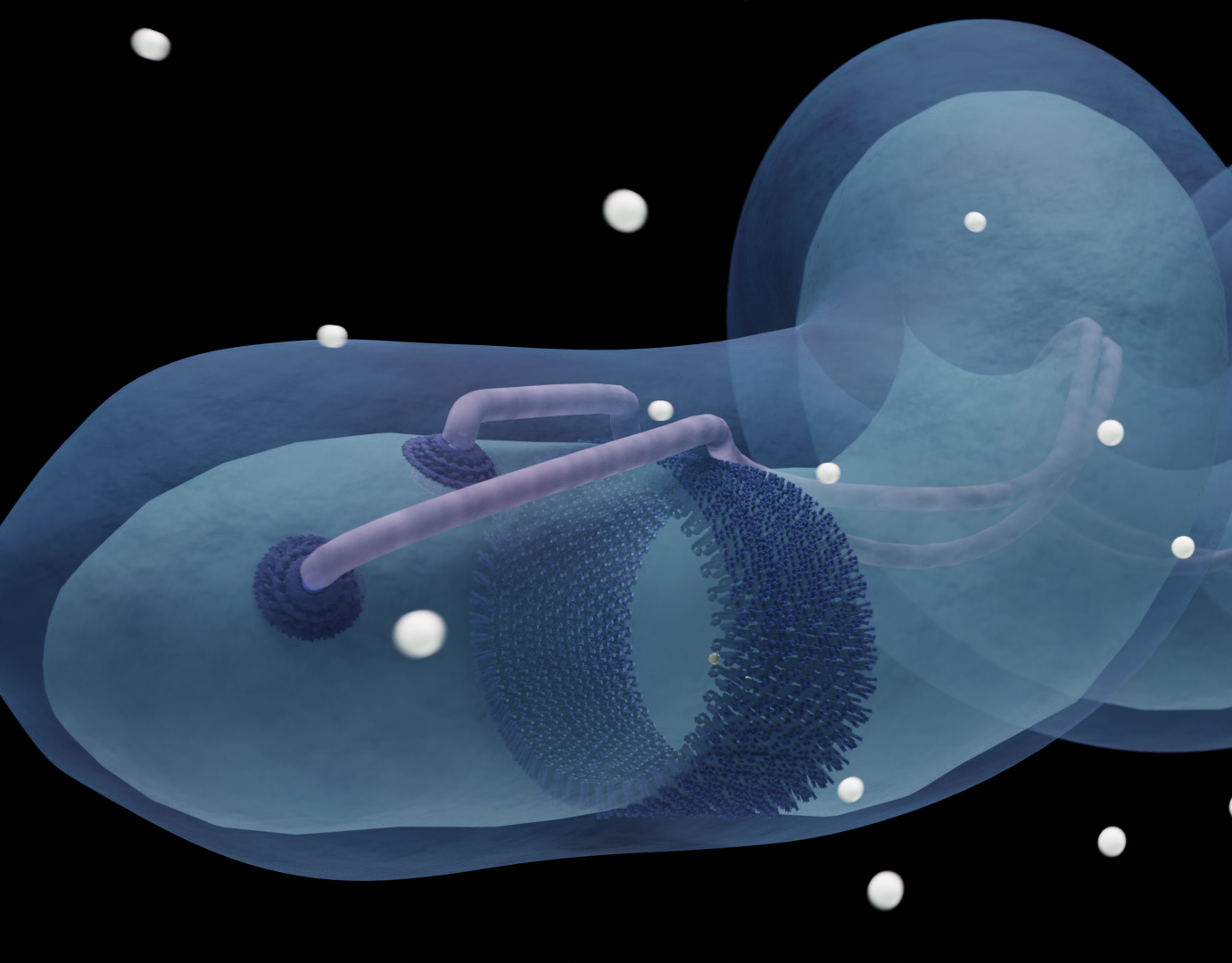
We are going full circle with this animation! My first finished science animation was about the flexible kinase domains in the chemoreceptorarray in E. coli (which was a part of my graduation project for my master thesis in 2020). Now, 5 years later, I have made an updated version of this animation! Although it still needs to be tweaked a bit, the concept is still the same: a demonstration of how the flexible kinase domains move when the receptors are activated. But I added more context in this version, by showing an overview of the actual array, and added more scientific data! For the first animation, I had to build the kinase domains by hand as the models were not available at the time, but now those structures have been uncovered.
This work is based on the research by Dr. Alise Muok, who has been a great help in instructing the movement. I placed the models in their positions with the illustrations from the paper by Cassidy et. al. The models used in this animation are from the Protein Database (2LP4).
Except where otherwise noted this work is protected under https://creativecommons.org/licenses/by-nc-nd/4.0/
References:
Cassidy CK, Qin Z, Frosio T, Gosink K, Yang Z, Sansom MSP, Stansfeld PJ, Parkinson JS, Zhang P. Structure of the native chemotaxis core signaling unit from phage E-protein lysed E. coli cells. mBio. 2023 Oct 31;14(5):e0079323. doi: 10.1128/mbio.00793-23. Epub 2023 Sep 29. PMID: 37772839; PMCID: PMC10653900.